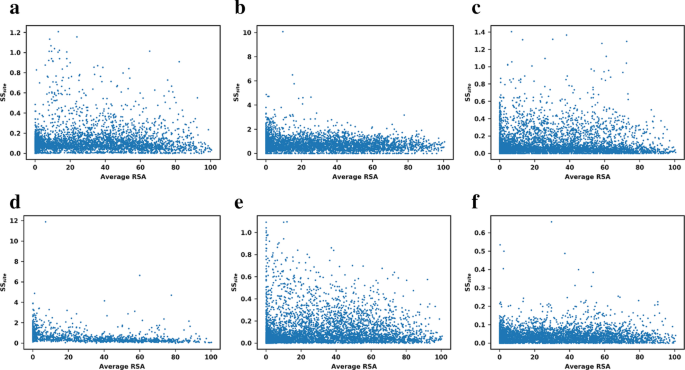
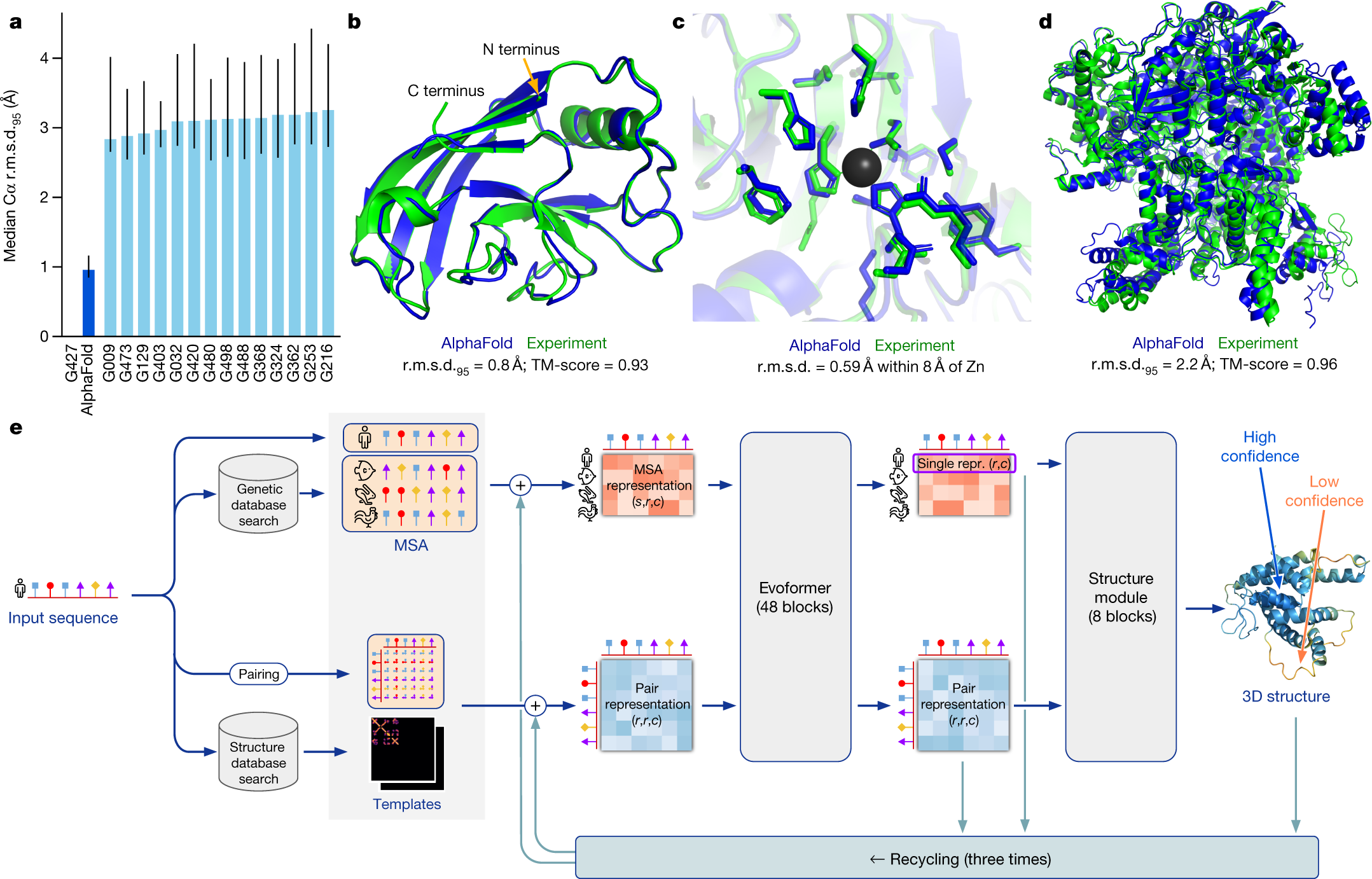
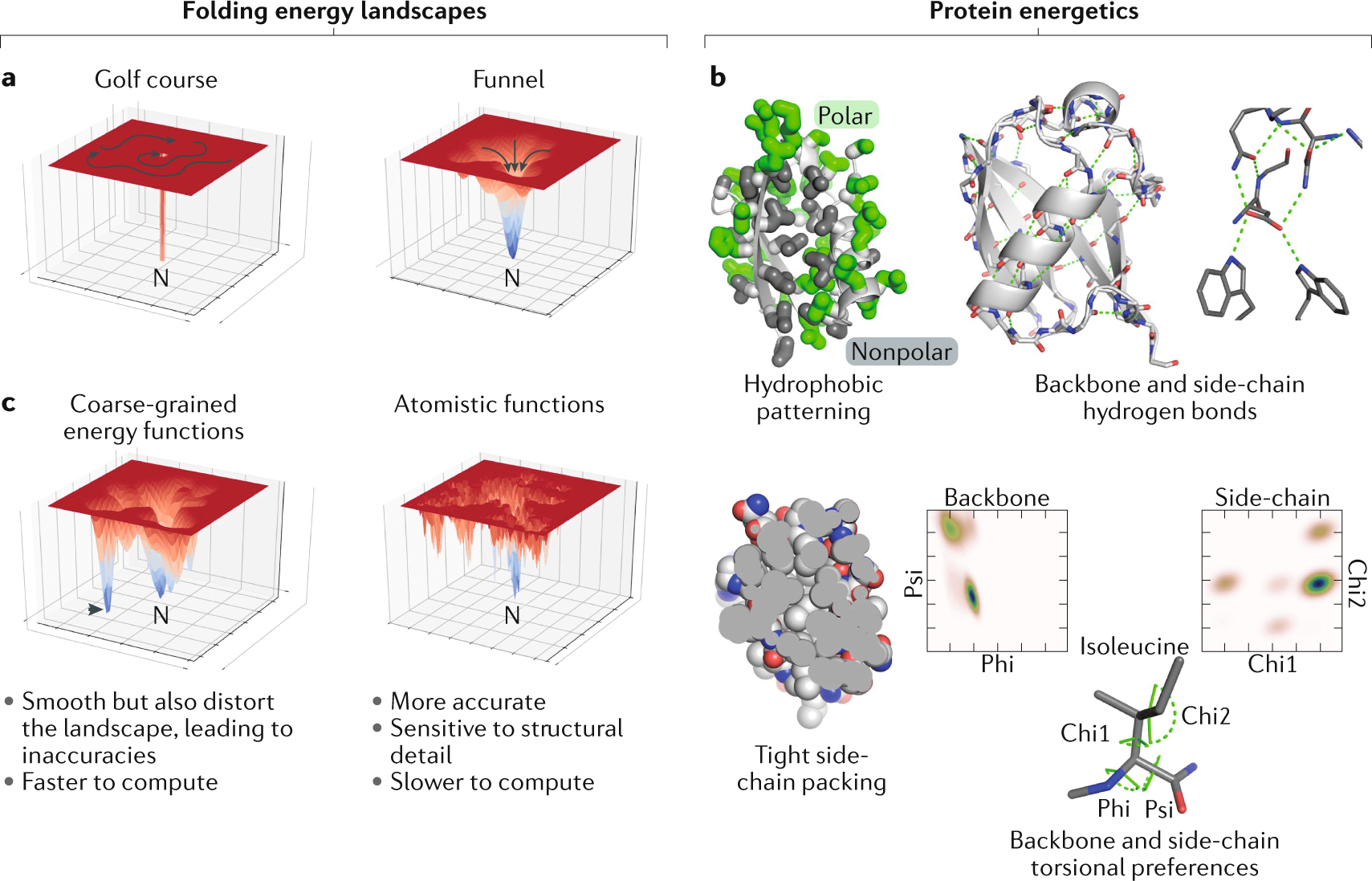
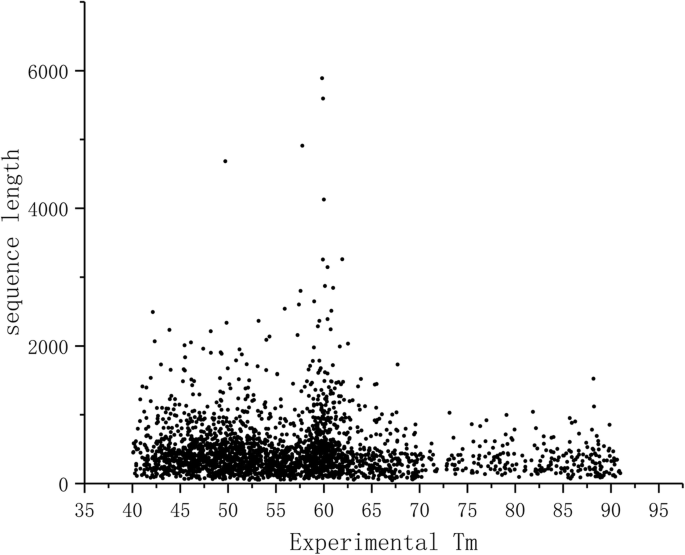
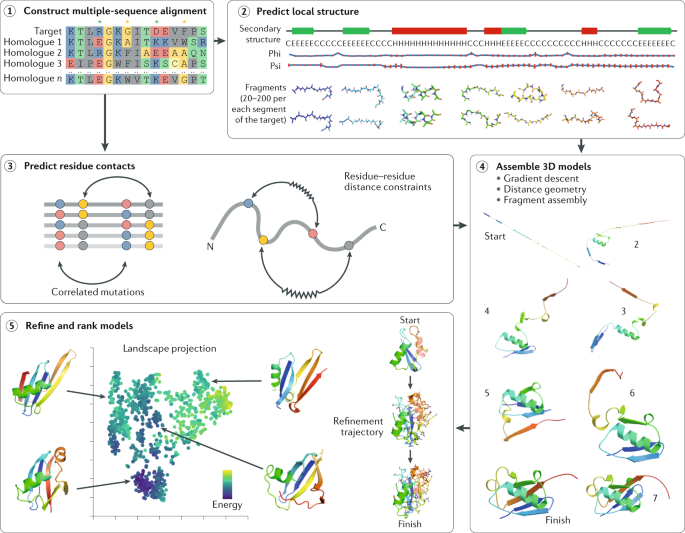
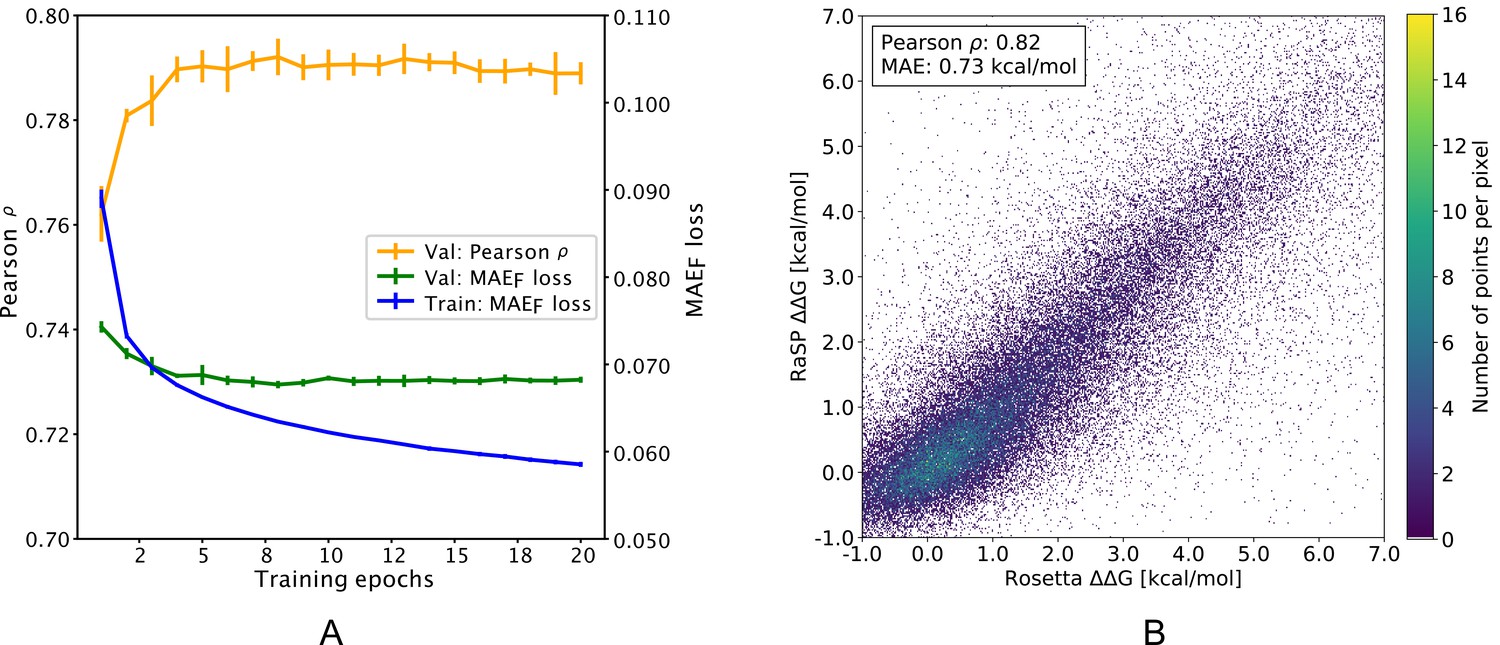
Predicting changes in protein thermodynamic stability upon point mutation with deep 3D convolutional neural networks | PLOS Computational Biology
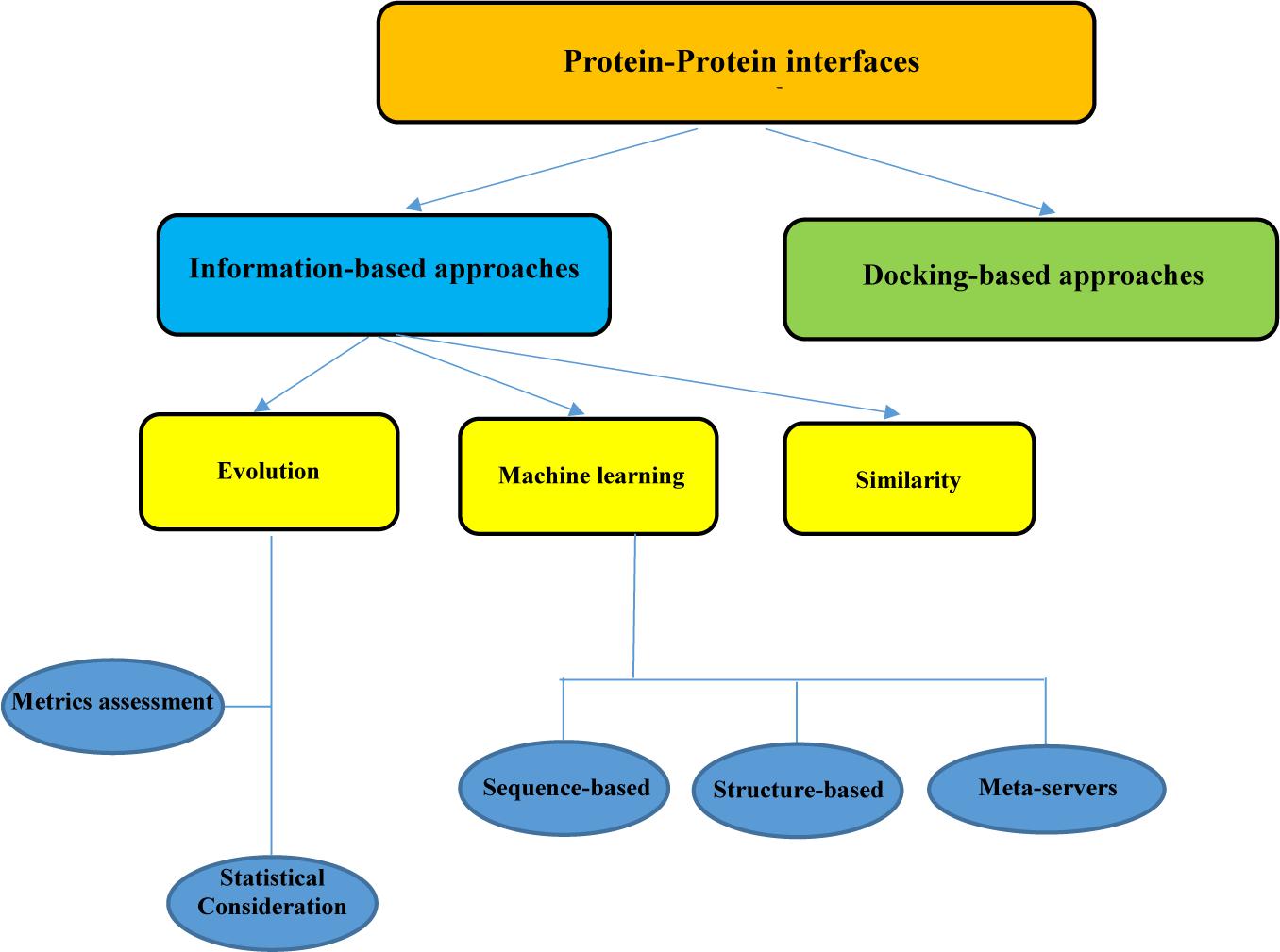
Frontiers | In silico Approaches for the Design and Optimization of Interfering Peptides Against Protein–Protein Interactions

Dissecting the stability determinants of a challenging de novo protein fold using massively parallel design and experimentation | PNAS

Computational Modeling of Protein Stability: Quantitative Analysis Reveals Solutions to Pervasive Problems - ScienceDirect

PROST: AlphaFold2-aware Sequence-Based Predictor to Estimate Protein Stability Changes upon Missense Mutations | Journal of Chemical Information and Modeling

Limitations and challenges in protein stability prediction upon genome variations: towards future applications in precision medicine - ScienceDirect

Predicting and interpreting large-scale mutagenesis data using analyses of protein stability and conservation - ScienceDirect

MPTherm-pred: Analysis and Prediction of Thermal Stability Changes upon Mutations in Transmembrane Proteins - ScienceDirect

Finding the ΔΔG spot: Are predictors of binding affinity changes upon mutations in protein–protein interactions ready for it? - Geng - 2019 - WIREs Computational Molecular Science - Wiley Online Library
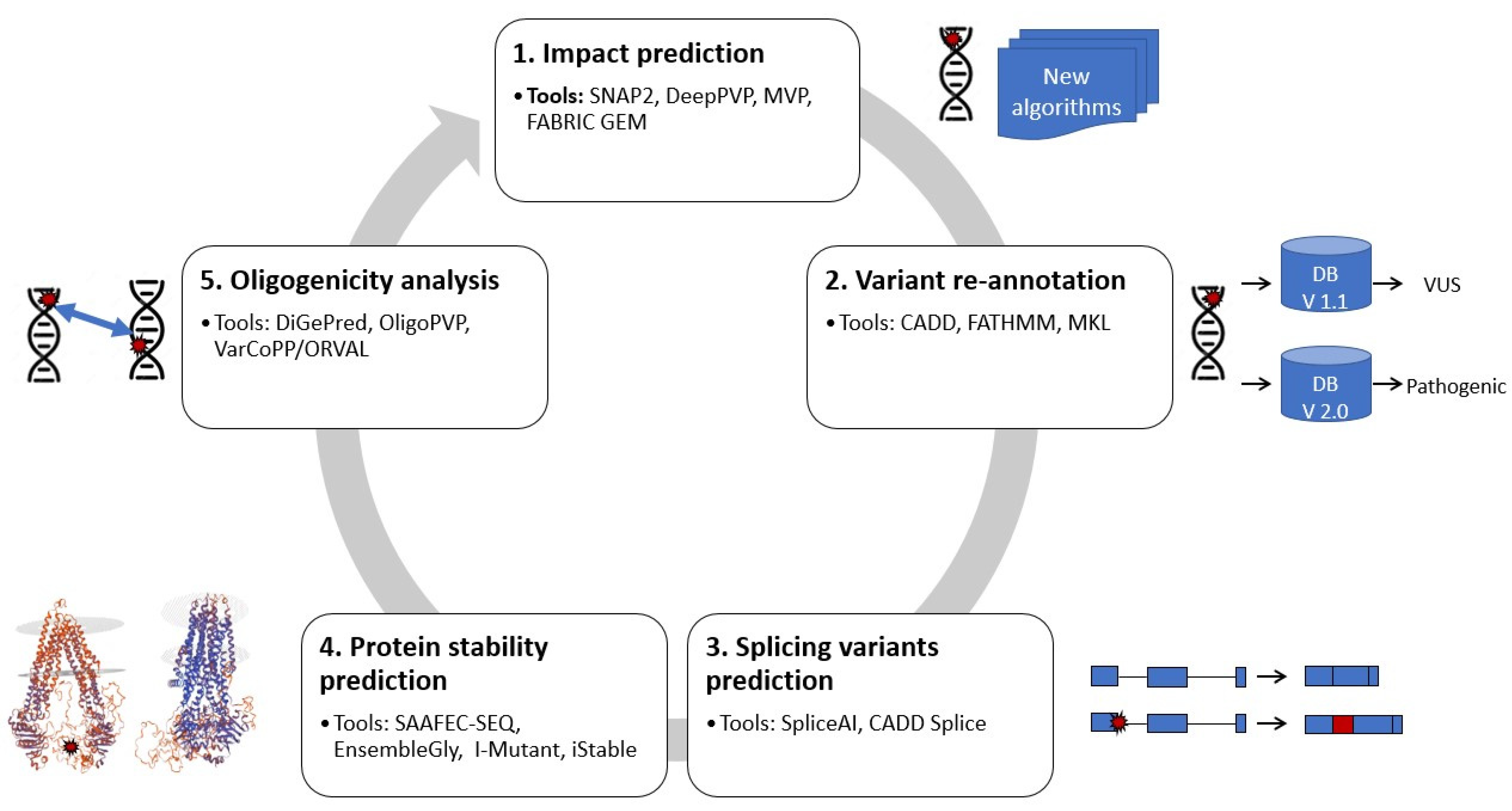
IJMS | Free Full-Text | New Developments and Possibilities in Reanalysis and Reinterpretation of Whole Exome Sequencing Datasets for Unsolved Rare Diseases Using Machine Learning Approaches
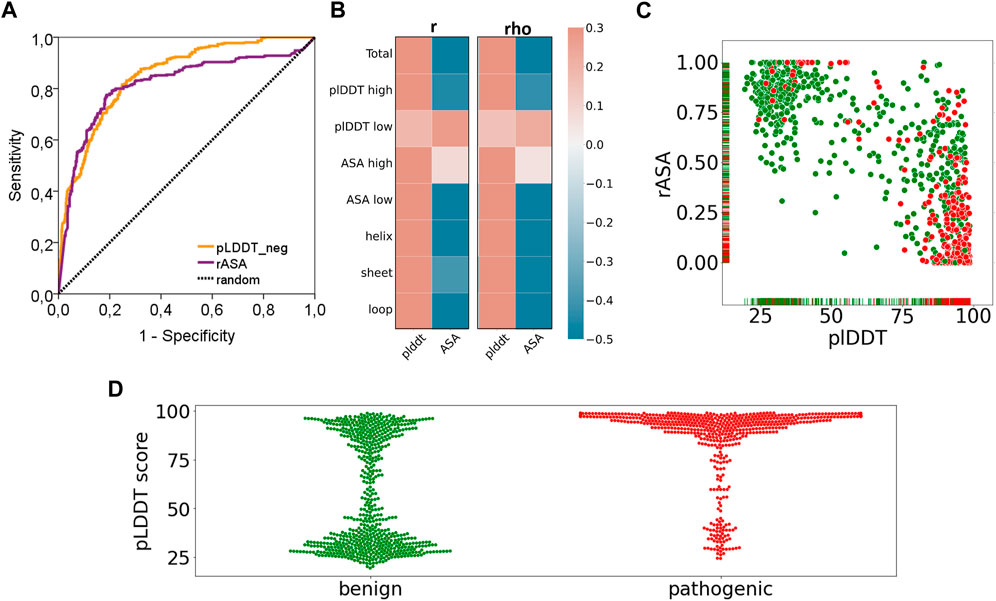
Frontiers | Evaluation of AlphaFold structure-based protein stability prediction on missense variations in cancer
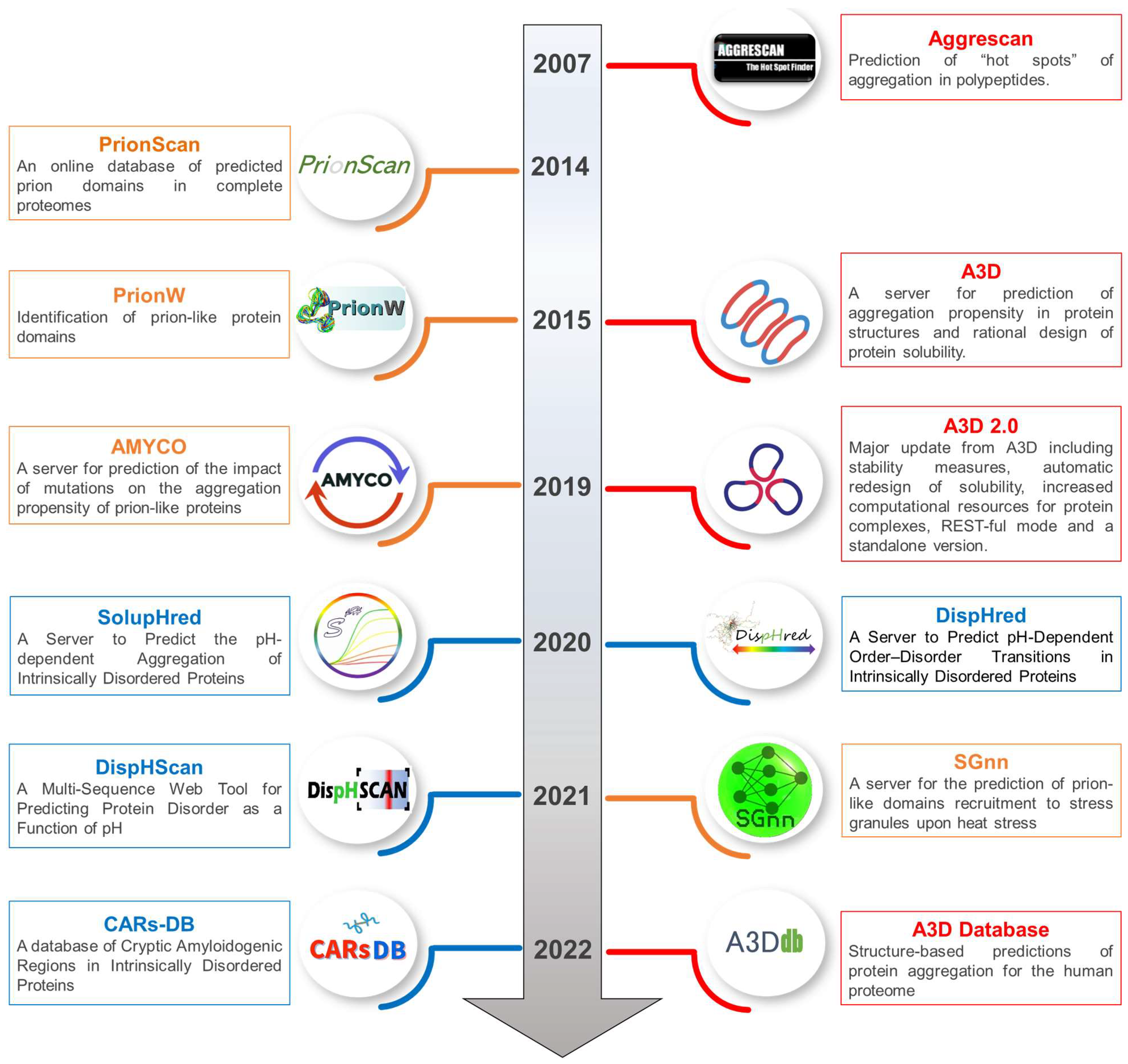
Biophysica | Free Full-Text | A Review of Fifteen Years Developing Computational Tools to Study Protein Aggregation

DynaMut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations - Rodrigues - 2021 - Protein Science - Wiley Online Library
PremPS: Predicting the impact of missense mutations on protein stability | PLOS Computational Biology